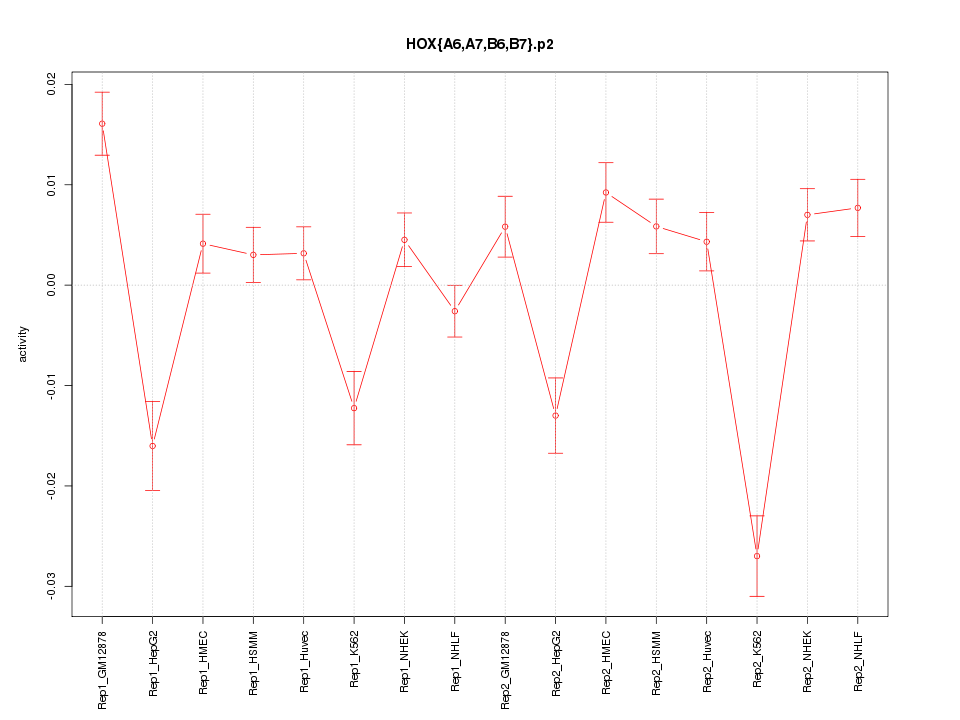
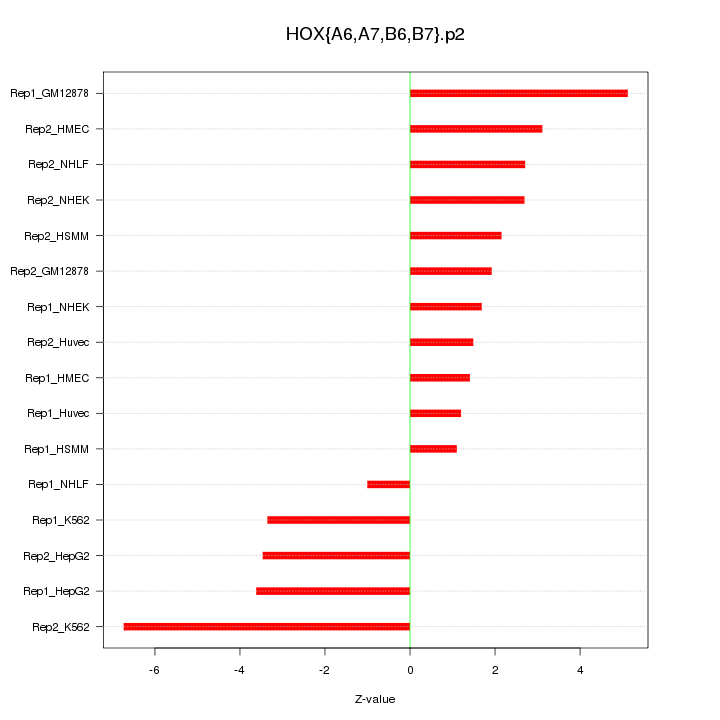
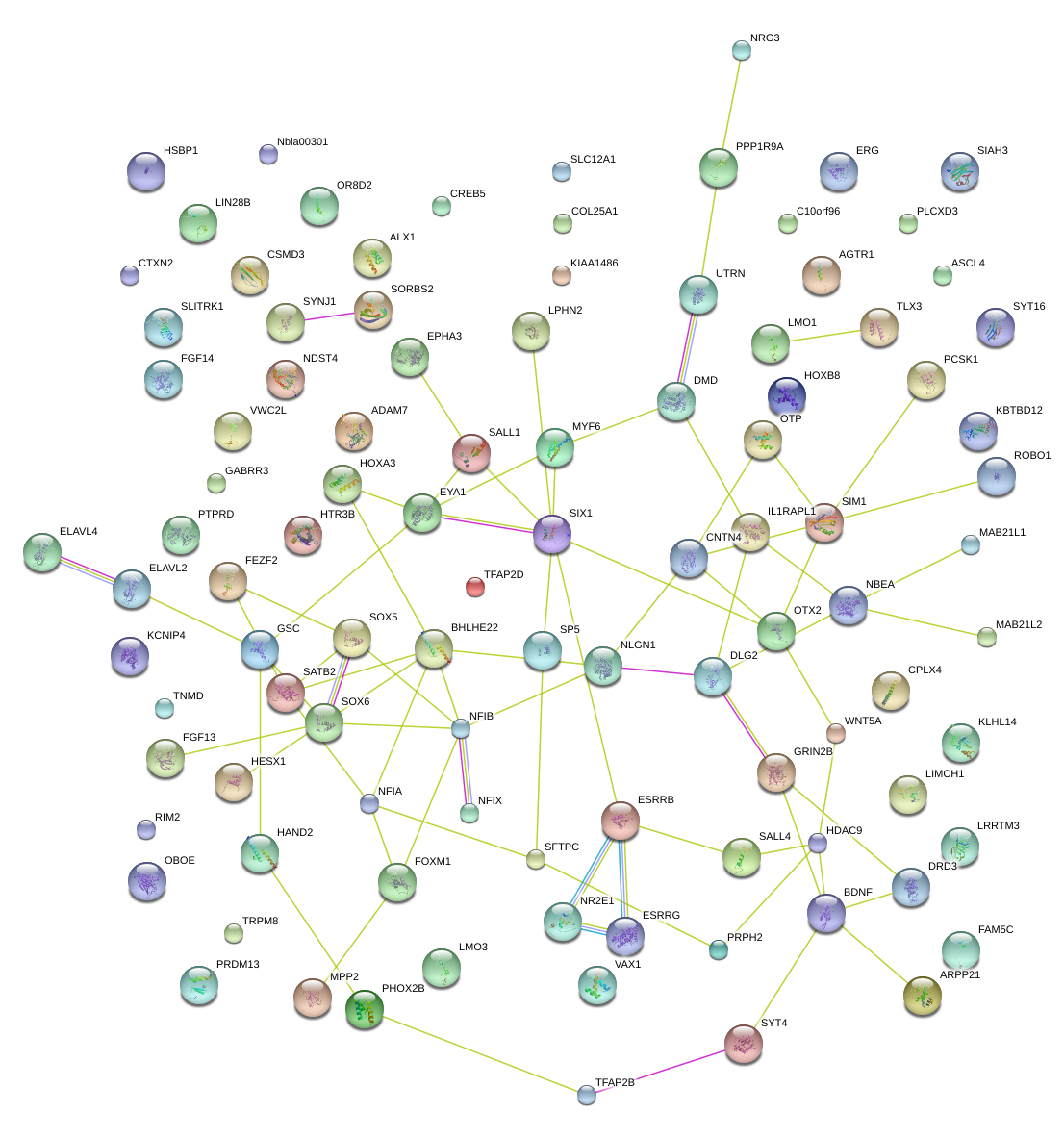
|
chr4_-_41579325
|
13.923
|
|
|
|
|
chr7_-_96471544
|
12.638
|
|
DLX6-AS1
|
DLX6 antisense RNA 1 (non-protein coding)
|
|
chr6_+_105511615
|
10.361
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr8_-_114518194
|
9.889
|
NM_052900
NM_198123
|
CSMD3
|
CUB and Sushi multiple domains 3
|
|
chr1_-_214963279
|
8.227
|
NM_001438
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr5_+_170668892
|
8.176
|
NM_021025
|
TLX3
|
T-cell leukemia homeobox 3
|
|
chr18_-_28606839
|
7.504
|
NM_020805
|
KLHL14
|
kelch-like 14 (Drosophila)
|
|
chr14_+_75907442
|
6.993
|
NM_004452
|
ESRRB
|
estrogen-related receptor beta
|
|
chr2_-_200033445
|
6.891
|
|
SATB2
|
SATB homeobox 2
|
|
chr13_-_45323757
|
6.649
|
NM_198849
|
SIAH3
|
seven in absentia homolog 3 (Drosophila)
|
|
chr10_+_83627422
|
6.573
|
NM_001165973
|
NRG3
|
neuregulin 3
|
|
chr1_-_188713324
|
6.371
|
NM_199051
|
FAM5C
|
family with sequence similarity 5, member C
|
|
chr14_-_94306104
|
6.249
|
NM_173849
|
GSC
|
goosecoid homeobox
|
|
chr13_-_101852028
|
5.974
|
NM_175929
|
FGF14
|
fibroblast growth factor 14
|
|
chr16_-_49742651
|
5.922
|
NM_002968
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr12_+_79625538
|
5.828
|
NM_002469
|
MYF6
|
myogenic factor 6 (herculin)
|
|
chr12_-_14024187
|
5.772
|
NM_000834
|
GRIN2B
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B
|
|
chr10_+_68355797
|
5.766
|
NM_178011
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3
|
|
chr12_-_16652202
|
5.742
|
NM_001001395
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr14_+_61532293
|
5.579
|
NM_031914
|
SYT16
|
synaptotagmin XVI
|
|
chr12_-_23995220
|
5.548
|
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr18_-_39111243
|
5.428
|
NM_020783
|
SYT4
|
synaptotagmin IV
|
|
chr4_-_110443247
|
5.316
|
NM_032518
NM_198721
|
COL25A1
|
collagen, type XXV, alpha 1
|
|
chr12_-_23995109
|
5.285
|
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr11_-_27679617
|
5.245
|
NM_170733
|
BDNF
|
brain-derived neurotrophic factor
|
|
chrX_+_99726445
|
5.200
|
NM_022144
|
TNMD
|
tenomodulin
|
|
chr11_-_27698800
|
5.200
|
NM_001143805
NM_001143806
NM_170732
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr11_-_16380967
|
5.052
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr3_+_174598937
|
5.035
|
NM_014932
|
NLGN1
|
neuroligin 1
|
|
chr1_+_61642287
|
4.907
|
|
NFIA
|
nuclear factor I/A
|
|
chr2_+_171280182
|
4.883
|
|
|
|
|
chr8_+_104900591
|
4.878
|
NM_014677
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr14_-_56346849
|
4.872
|
NM_021728
|
OTX2
|
orthodenticle homeobox 2
|
|
chr3_-_62334229
|
4.866
|
NM_018008
|
FEZF2
|
FEZ family zinc finger 2
|
|
chrX_+_28515479
|
4.852
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr21_-_38955487
|
4.850
|
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr18_-_55136673
|
4.837
|
NM_181654
|
CPLX4
|
complexin 4
|
|
chr6_+_50894397
|
4.783
|
NM_003221
|
TFAP2B
|
transcription factor AP-2 beta (activating enhancer binding protein 2 beta)
|
|
chrX_-_137649180
|
4.780
|
NM_033642
|
FGF13
|
fibroblast growth factor 13
|
|
chr3_+_35696119
|
4.701
|
NM_001025068
NM_001025069
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr4_-_116254381
|
4.680
|
NM_022569
|
NDST4
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4
|
|
chr9_-_8723893
|
4.649
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr11_-_84311867
|
4.588
|
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr2_+_225973845
|
4.582
|
NM_020864
|
KIAA1486
|
KIAA1486
|
|
chr5_-_76970129
|
4.572
|
|
OTP
|
orthopedia homeobox
|
|
chr2_+_171280046
|
4.542
|
NM_001003845
|
SP5
|
Sp5 transcription factor
|
|
chr7_+_28418668
|
4.498
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr13_-_34948758
|
4.498
|
NM_005584
|
MAB21L1
|
mab-21-like 1 (C. elegans)
|
|
chr13_-_83354474
|
4.458
|
NM_052910
|
SLITRK1
|
SLIT and NTRK-like family, member 1
|
|
chr2_+_234490781
|
4.381
|
NM_024080
|
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8
|
|
chr11_-_123695302
|
4.242
|
NM_001002918
|
OR8D2
|
olfactory receptor, family 8, subfamily D, member 2
|
|
chr6_-_101018271
|
4.122
|
NM_005068
|
SIM1
|
single-minded homolog 1 (Drosophila)
|
|
chr5_-_76970264
|
4.105
|
NM_032109
|
OTP
|
orthopedia homeobox
|
|
chr5_-_95794416
|
4.067
|
NM_000439
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr4_+_174688183
|
4.045
|
|
NBLA00301
|
Nbla00301
|
|
chr13_-_83354425
|
3.994
|
|
SLITRK1
|
SLIT and NTRK-like family, member 1
|
|
chr11_+_113280727
|
3.990
|
NM_006028
|
HTR3B
|
5-hydroxytryptamine (serotonin) receptor 3B
|
|
chr4_-_41445693
|
3.951
|
NM_003924
|
PHOX2B
|
paired-like homeobox 2b
|
|
chr12_+_84198015
|
3.896
|
NM_006982
|
ALX1
|
ALX homeobox 1
|
|
chr8_+_65655367
|
3.875
|
NM_152414
|
BHLHE22
|
basic helix-loop-helix family, member e22
|
|
chr6_+_108593907
|
3.845
|
NM_003269
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1
|
|
chr10_+_118073929
|
3.841
|
NM_198515
|
C10orf96
|
chromosome 10 open reading frame 96
|
|
chr12_+_106692291
|
3.829
|
NM_203436
|
ASCL4
|
achaete-scute complex homolog 4 (Drosophila)
|
|
chr4_-_187114407
|
3.817
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr4_-_174687931
|
3.778
|
NM_021973
|
HAND2
|
heart and neural crest derivatives expressed 2
|
|
chr3_-_55489973
|
3.737
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr15_+_46285789
|
3.693
|
NM_000338
NM_001184832
|
SLC12A1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 1
|
|
chr20_-_49852353
|
3.651
|
NM_020436
|
SALL4
|
sal-like 4 (Drosophila)
|
|
chr1_+_81544432
|
3.637
|
|
LPHN2
|
latrophilin 2
|
|
chr3_-_79899748
|
3.632
|
NM_002941
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr15_+_46271158
|
3.604
|
NM_001145668
|
CTXN2
SLC12A1
|
cortexin 2
solute carrier family 12 (sodium/potassium/chloride transporters), member 1
|
|
chr6_+_50789215
|
3.595
|
NM_172238
|
TFAP2D
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta)
|
|
chr13_+_34948977
|
3.588
|
|
NBEA
|
neurobeachin
|
|
chr17_-_44047299
|
3.586
|
NM_024016
|
HOXB8
|
homeobox B8
|
|
chr7_+_94377145
|
3.569
|
NM_001166161
NM_001166162
NM_001166163
|
PPP1R9A
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A
|
|
chr7_-_27133163
|
3.546
|
NM_153631
|
HOXA3
|
homeobox A3
|
|
chr12_+_63510481
|
3.539
|
|
|
|
|
chr13_+_34948885
|
3.446
|
|
NBEA
|
neurobeachin
|
|
chr8_+_24354387
|
3.445
|
NM_003817
|
ADAM7
|
ADAM metallopeptidase domain 7
|
|
chr3_+_35697432
|
3.436
|
NM_198399
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr6_+_100161370
|
3.418
|
NM_021620
|
PRDM13
|
PR domain containing 13
|
|
chr7_+_18296569
|
3.413
|
|
HDAC9
|
histone deacetylase 9
|
|
chr3_+_89239494
|
3.399
|
|
EPHA3
|
EPH receptor A3
|
|
chr4_-_21308415
|
3.325
|
NM_001035003
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr14_-_60185659
|
3.314
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr4_-_20914626
|
3.286
|
NM_147183
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr3_-_57209319
|
3.257
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr1_+_50348052
|
3.244
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr3_-_99236520
|
3.217
|
NM_001105580
|
GABRR3
|
gamma-aminobutyric acid (GABA) receptor, rho 3
|
|
chr4_-_41445479
|
3.196
|
|
PHOX2B
|
paired-like homeobox 2b
|
|
chr8_-_72431274
|
3.184
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr10_-_118887801
|
3.182
|
NM_001112704
NM_199131
|
VAX1
|
ventral anterior homeobox 1
|
|
chr3_+_129124591
|
3.139
|
NM_207335
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12
|
|
chr4_+_41309690
|
3.083
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr3_+_149930656
|
3.054
|
NM_032049
|
AGTR1
|
angiotensin II receptor, type 1
|
|
chr2_+_214984705
|
3.033
|
NM_001080500
|
VWC2L
|
von Willebrand factor C domain containing protein 2-like
|
|
chr3_-_115380539
|
3.011
|
NM_000796
NM_033663
|
DRD3
|
dopamine receptor D3
|
|
chr3_+_2528272
|
3.010
|
|
CNTN4
|
contactin 4
|
|
chr1_+_50344550
|
3.006
|
NM_001144775
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chrX_-_33267646
|
2.934
|
NM_000109
|
DMD
|
dystrophin
|
|
chr5_-_41546384
|
2.916
|
NM_001005473
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3
|
|
chr4_+_41309482
|
2.901
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr5_-_22248630
|
2.889
|
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2)
|
|
chr1_+_175406685
|
2.886
|
|
FAM5B
|
family with sequence similarity 5, member B
|
|
chr8_-_93176819
|
2.867
|
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr18_-_29882555
|
2.848
|
NM_001198549
|
NOL4
|
nucleolar protein 4
|
|
chr4_-_187114863
|
2.837
|
NM_021069
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr18_-_3864252
|
2.821
|
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr17_+_72047709
|
2.807
|
NM_001077620
|
PRCD
|
progressive rod-cone degeneration
|
|
chr4_-_20594645
|
2.798
|
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr12_-_23993830
|
2.777
|
NM_006940
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr11_-_36576352
|
2.767
|
NM_000536
|
RAG2
|
recombination activating gene 2
|
|
chr12_-_16649211
|
2.764
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr4_-_184480572
|
2.763
|
NM_001185149
|
CLDN24
|
claudin 24
|
|
chr1_+_50347176
|
2.761
|
NM_001144774
NM_021952
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr18_+_57308754
|
2.761
|
NM_031891
|
CDH20
|
cadherin 20, type 2
|
|
chrX_+_11221453
|
2.757
|
NM_001142
NM_182680
NM_182681
|
AMELX
|
amelogenin, X-linked
|
|
chr1_+_50342168
|
2.752
|
NM_001144776
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr13_-_52320774
|
2.751
|
NM_002590
NM_032949
|
PCDH8
|
protocadherin 8
|
|
chr11_-_27679175
|
2.726
|
NM_001143808
NM_001143809
NM_001143810
NM_001143811
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr1_+_65503011
|
2.722
|
NM_014787
|
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr6_-_94185780
|
2.718
|
NM_004440
|
EPHA7
|
EPH receptor A7
|
|
chr15_+_56217686
|
2.706
|
NM_020980
|
AQP9
|
aquaporin 9
|
|
chr12_-_16321885
|
2.682
|
NM_001170798
|
SLC15A5
|
solute carrier family 15, member 5
|
|
chr11_-_84312085
|
2.673
|
NM_001364
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr5_-_88004891
|
2.660
|
|
LOC645323
|
hypothetical LOC645323
|
|
chr3_+_35658852
|
2.659
|
NM_016300
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr5_-_58918031
|
2.657
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr7_-_19151509
|
2.645
|
NM_152898
|
FERD3L
|
Fer3-like (Drosophila)
|
|
chr14_-_56342097
|
2.644
|
NM_172337
|
OTX2
|
orthodenticle homeobox 2
|
|
chr4_+_41309668
|
2.644
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr11_+_22316242
|
2.631
|
NM_020346
|
SLC17A6
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6
|
|
chr18_+_30427279
|
2.621
|
NM_001128175
NM_001198938
NM_001198939
NM_001198940
|
DTNA
|
dystrobrevin, alpha
|
|
chr9_-_72673753
|
2.610
|
NM_001007470
NM_020952
NM_024971
NM_206944
NM_206945
NM_206946
NM_206947
NM_206948
|
TRPM3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr7_-_31347032
|
2.608
|
NM_022728
|
NEUROD6
|
neurogenic differentiation 6
|
|
chrX_+_9391314
|
2.602
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr11_-_27700180
|
2.591
|
NM_170731
|
BDNF
|
brain-derived neurotrophic factor
|
|
chrX_-_32340291
|
2.589
|
NM_004011
NM_004012
|
DMD
|
dystrophin
|
|
chr3_+_68136056
|
2.581
|
NM_213609
|
FAM19A1
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A1
|
|
chr7_+_18501876
|
2.555
|
NM_014707
NM_178423
|
HDAC9
|
histone deacetylase 9
|
|
chr1_+_168898898
|
2.552
|
|
PRRX1
|
paired related homeobox 1
|
|
chr14_-_80934523
|
2.539
|
NM_033104
|
STON2
|
stonin 2
|
|
chrX_+_135558001
|
2.532
|
NM_000074
|
CD40LG
|
CD40 ligand
|
|
chr4_+_114190233
|
2.524
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr3_+_1109619
|
2.521
|
NM_014461
|
CNTN6
|
contactin 6
|
|
chr11_-_30558495
|
2.513
|
NM_001584
|
MPPED2
|
metallophosphoesterase domain containing 2
|
|
chr2_+_211166323
|
2.512
|
NM_001122634
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial
|
|
chr1_+_50347188
|
2.503
|
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr7_+_113842470
|
2.503
|
|
FOXP2
|
forkhead box P2
|
|
chr8_-_93176618
|
2.490
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr3_-_139333813
|
2.486
|
NM_016161
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase
|
|
chr1_-_195302973
|
2.464
|
NM_001994
|
F13B
|
coagulation factor XIII, B polypeptide
|
|
chr7_-_83116414
|
2.459
|
NM_012431
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E
|
|
chr4_+_119174947
|
2.453
|
NM_004784
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3
|
|
chr12_+_40117751
|
2.452
|
NM_013377
|
PDZRN4
|
PDZ domain containing ring finger 4
|
|
chrX_+_82649924
|
2.448
|
NM_000307
|
POU3F4
|
POU class 3 homeobox 4
|
|
chr7_-_36992221
|
2.443
|
NM_130442
|
ELMO1
|
engulfment and cell motility 1
|
|
chr14_-_57030227
|
2.414
|
NM_018168
|
C14orf105
|
chromosome 14 open reading frame 105
|
|
chr1_-_93030538
|
2.402
|
NM_005665
|
EVI5
|
ecotropic viral integration site 5
|
|
chr12_-_10846435
|
2.394
|
NM_023919
|
TAS2R7
|
taste receptor, type 2, member 7
|
|
chr7_+_113842407
|
2.392
|
|
FOXP2
|
forkhead box P2
|
|
chr5_-_1935764
|
2.390
|
NM_016358
|
IRX4
|
iroquois homeobox 4
|
|
chr2_+_137464931
|
2.383
|
NM_001080427
|
THSD7B
|
thrombospondin, type I, domain containing 7B
|
|
chr11_-_26700121
|
2.339
|
NM_178498
|
SLC5A12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12
|
|
chr18_+_50409387
|
2.337
|
NM_173629
|
C18orf26
|
chromosome 18 open reading frame 26
|
|
chr18_+_53170718
|
2.335
|
NM_015879
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3
|
|
chr1_-_163680941
|
2.328
|
NM_006917
|
RXRG
|
retinoid X receptor, gamma
|
|
chr3_+_116824840
|
2.319
|
NM_001130064
NM_002045
|
GAP43
|
growth associated protein 43
|
|
chr12_+_84792203
|
2.276
|
NM_006183
|
NTS
|
neurotensin
|
|
chr12_-_16650697
|
2.275
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr8_-_91164100
|
2.273
|
NM_004929
|
CALB1
|
calbindin 1, 28kDa
|
|
chr4_-_44348414
|
2.273
|
NM_182592
|
YIPF7
|
Yip1 domain family, member 7
|
|
chr5_-_145232723
|
2.247
|
NM_001080516
|
GRXCR2
|
glutaredoxin, cysteine rich 2
|
|
chr18_-_68683913
|
2.246
|
NM_138999
NM_153181
|
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1
|
|
chr13_+_77213295
|
2.230
|
NM_144595
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chr4_+_41395493
|
2.224
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr12_+_52697160
|
2.214
|
|
HOXC5
HOXC6
|
homeobox C5
homeobox C6
|
|
chr2_-_183095308
|
2.213
|
NM_005019
NM_001003683
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent
|
|
chr6_-_64087806
|
2.205
|
NM_001143940
NM_016571
|
LGSN
|
lengsin, lens protein with glutamine synthetase domain
|
|
chr5_-_134899537
|
2.195
|
NM_006161
|
NEUROG1
|
neurogenin 1
|
|
chr3_-_54937001
|
2.189
|
NM_020678
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1
|
|
chr2_-_182253457
|
2.186
|
NM_002500
|
NEUROD1
|
neurogenic differentiation 1
|
|
chr12_+_79362256
|
2.183
|
NM_001145026
|
PTPRQ
|
protein tyrosine phosphatase, receptor type, Q
|
|
chr4_-_164754106
|
2.182
|
NM_017923
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr17_-_30909219
|
2.178
|
NM_001129820
|
SLFN14
|
schlafen family member 14
|
|
chr18_-_30057512
|
2.169
|
NM_001198546
NM_001198548
NM_003787
|
NOL4
|
nucleolar protein 4
|
|
chr3_-_71262426
|
2.167
|
|
FOXP1
|
forkhead box P1
|
|
chr1_+_198263352
|
2.167
|
NM_003822
NM_205860
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2
|
|
chr7_+_28441758
|
2.164
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr12_-_16650687
|
2.146
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr14_-_100105883
|
2.140
|
NM_020836
|
BEGAIN
|
brain-enriched guanylate kinase-associated homolog (rat)
|
|
chr13_-_102517196
|
2.119
|
NM_000452
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 2
|
|
chr4_+_93444572
|
2.117
|
NM_001510
|
GRID2
|
glutamate receptor, ionotropic, delta 2
|
|
chr7_-_72676750
|
2.104
|
NM_032951
NM_032952
NM_032953
NM_032954
|
MLXIPL
|
MLX interacting protein-like
|
|
chr11_-_31782169
|
2.104
|
|
PAX6
|
paired box 6
|
|
chr18_+_74841262
|
2.099
|
NM_171999
|
SALL3
|
sal-like 3 (Drosophila)
|
|
chr4_+_114433551
|
2.076
|
|
ANK2
|
ankyrin 2, neuronal
|
|
chr11_-_128217550
|
2.056
|
NM_000220
|
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1
|
|
chr1_-_214663360
|
2.055
|
NM_007123
NM_206933
|
USH2A
|
Usher syndrome 2A (autosomal recessive, mild)
|
|
chr3_-_71376919
|
2.044
|
|
FOXP1
|
forkhead box P1
|